Tutorial
Searching the database
Currently, you can search using the gene Id (if you know) or any probably function for a gene. As an example, searching for the keyword “auxin” will result in 325 table entries having auxin as a keyword in their function. The resultant table will show the accession, chromosome number, Gene ID, Gene Ontology (GO), start and end position, and the DNA strand. Links have been provided in the table to view the fasta sequence of that particular gene, to view the gene in JBrowse2, and to view the dashboard summarizing the statistics of BLAST results of that gene with other wheat varieties (from a pan-genomic perspective). This dashboard provides various statistical information derived from the BLAST of T. monococcum genes against the other wheat varieties. The statistics provided here include the length of gene sequence, number of germplasm matched along with the individual genomes, length matched, scores etc.
JBrowse2
Previously released Jbrowse1 (Buels et al., 2016; Skinner, Uzilov, Stein, Mungall, & Holmes, 2009) had some limitations like only one genome would be displayed and needed more support for new software libraries. Here, we have used the linear genome viewer of JBrowse2 to show the genome assemblies of T. monococcum TA299 and T. monococcum TA10622. Both assemblies can be accessed by individual chromosomes. Gene model annotations were linked to each of the assemblies which include gene structure and putative function for both high-confidence and low-confidence genes models. However, a separate annotation file has been provided for only high-confidence genes for those who are interested in high-confidence genes. Annotated Transposable Elements (TE) were also associated with each of the genomes.
How to view the linear synteny in JBrowse2
Linear synteny has been provided between available genomes in the browser. Steps to view the linear synteny is as follows:
1. From the JBrowse2 home page click on Empty link (highlighted).
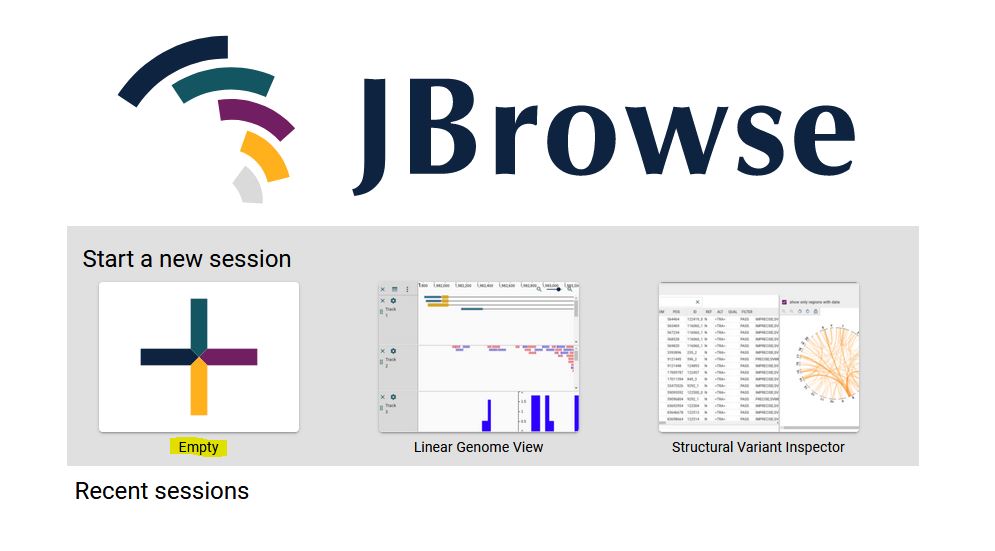
2. Select Linear synteny view from the dropdown menu and click ‘Launch view’.
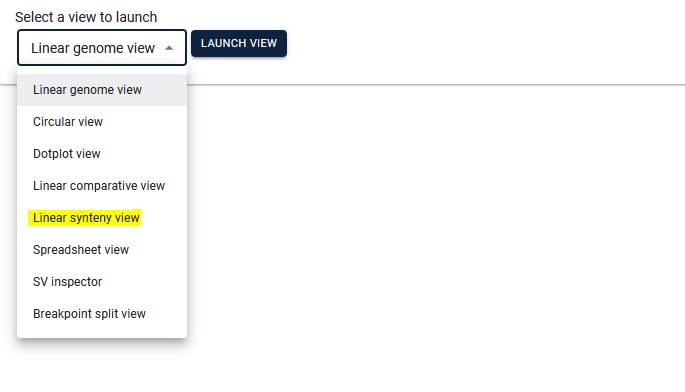
3. Select the two species for which you want to see synteny, then select the existing track (automatically the respective file will be displayed) and click Launch.

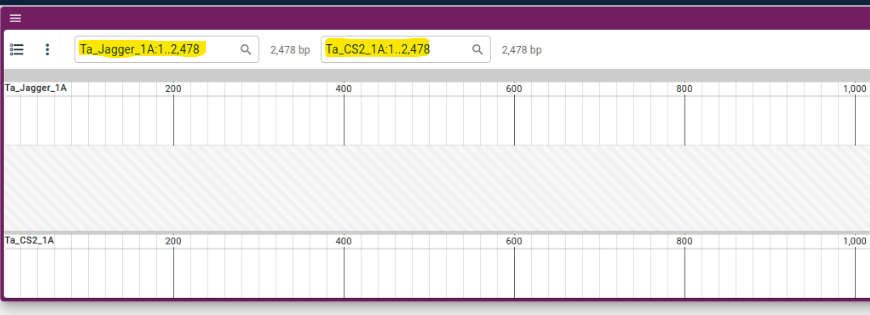
4. From the two dropdown menus, select the respective chromosomes (it will be best to choose the same chromosome number on both the genomes).
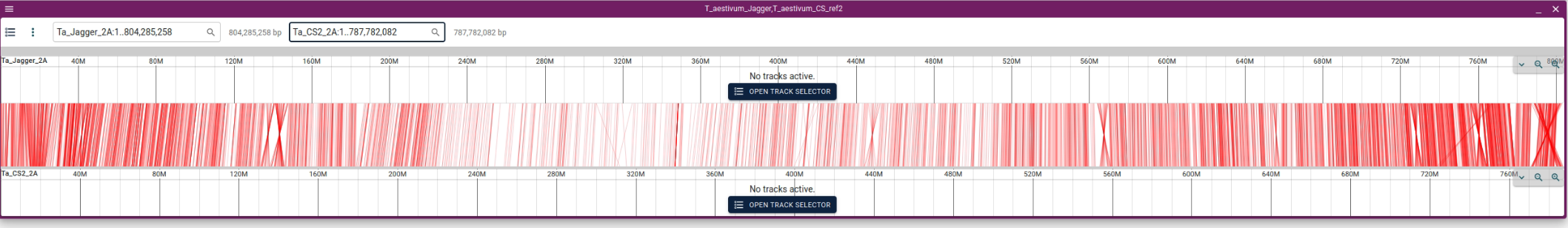
5. Synteny will be displayed.
SequenceServer2
Sequenceserver is a tool that provide BLAST facility. In this database, we have taken care of pan-genomic perspective of large genome and provided group wise BLAST databases. For example, if a user wants to do a BLAST search against only A genome of all the wheat varieties, he/she can select ‘Group A’ and easily do the search.
Kablammo
Kablammo accepts BLAST result output files in XML format and will illustrate exactly which portions of the query sequence mapped to which portions of the subject sequence. Kablammo takes an XML file as an input (which can be downloaded from SequenceServer output) and provides an interactive figure for each subject matched. The query as well as subject sequences can be zoomed-out or zoomed-in for better results viewing experience. Other features like image export, alignment viewer, and exporter are also available.