Currently, you can search using the gene Id (if you know) or any probably function for a gene. As an example, searching for the keyword “auxin” will result in 325 table entries having auxin as a keyword in their function. The resultant table will show the accession, chromosome number, Gene ID, Gene Ontology (GO), start and end position, and the DNA strand. Links have been provided in the table to view the fasta sequence of that particular gene, to view the gene in JBrowse2, and to view the dashboard summarizing the statistics of BLAST results of that gene with other wheat varieties (from a pan-genomic perspective). This dashboard provides various statistical information derived from the BLAST of T. monococcum genes against the other wheat varieties. The statistics provided here include the length of gene sequence, number of germplasm matched along with the individual genomes, length matched, scores etc.
Previously released Jbrowse1 (Buels et al., 2016; Skinner, Uzilov, Stein, Mungall, & Holmes, 2009) had some limitations like only one genome would be displayed and needed more support for new software libraries. Here, we have used the linear genome viewer of JBrowse2 to show the genome assemblies of T. monococcum TA299 and T. monococcum TA10622. Both assemblies can be accessed by individual chromosomes. Gene model annotations were linked to each of the assemblies which include gene structure and putative function for both high-confidence and low-confidence genes models. However, a separate annotation file has been provided for only high-confidence genes for those who are interested in high-confidence genes. Annotated Transposable Elements (TE) were also associated with each of the genomes.
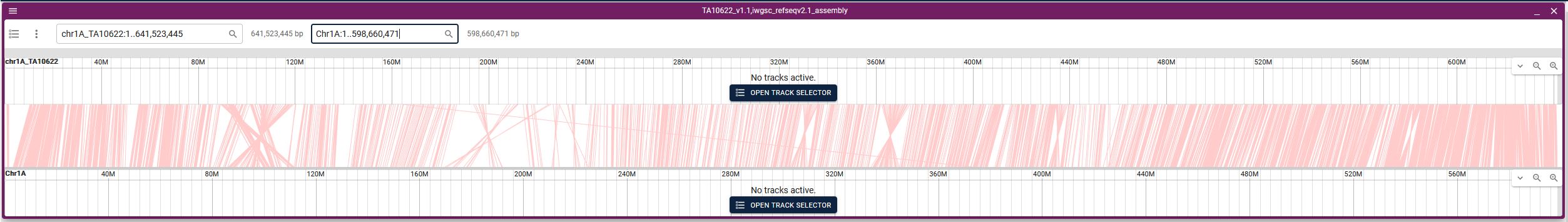
Linear synteny has been provided between available genomes in the browser. Steps to view the linear synteny is as follows:
1. From the JBrowse2 home page click on Empty link (highlighted).
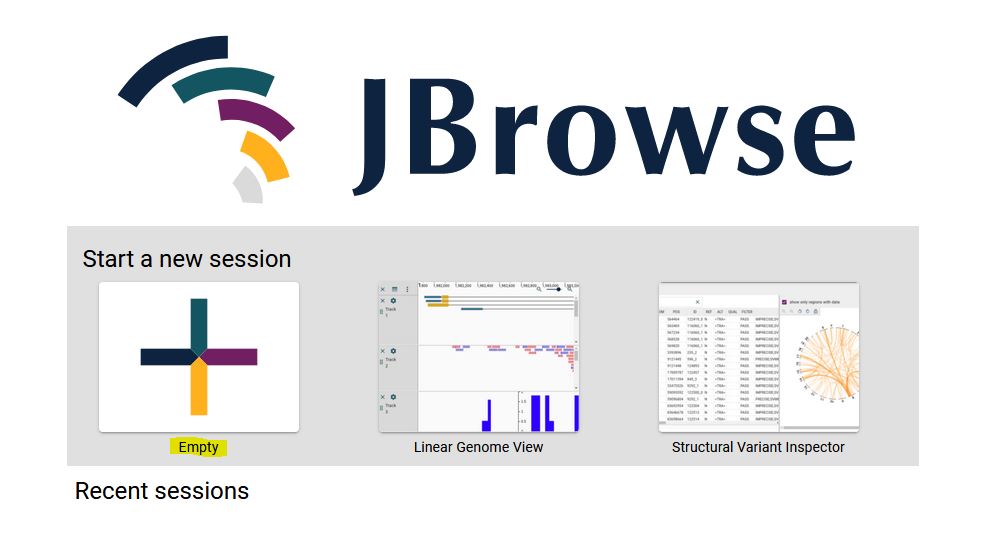
2. Select Linear synteny view from the dropdown menu and click ‘Launch view’.
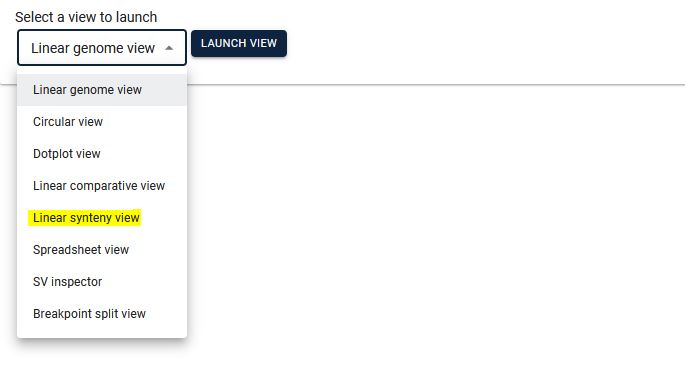
3. Select the two species for which you want to see synteny, then select the existing track (automatically the respective file will be displayed) and click Launch.
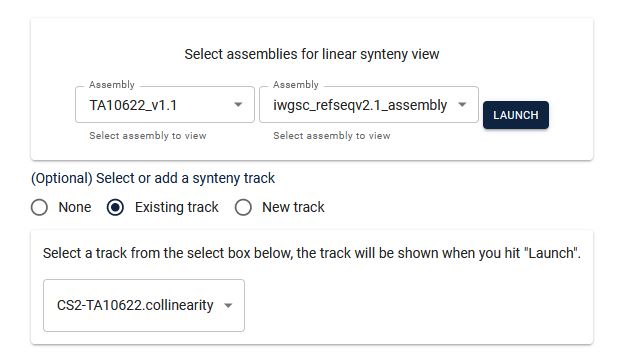
4. From the two dropdown menus, select the respective chromosomes (it will be best to choose the same chromosome number on both the genomes).
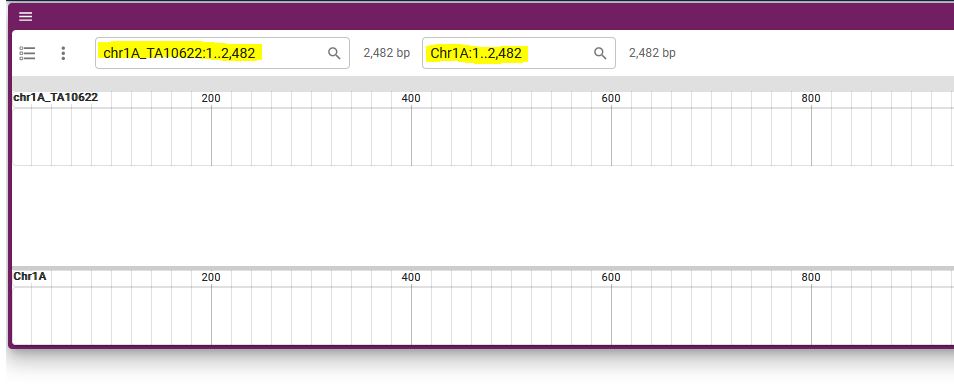
5. Synteny will be displayed.
Sequenceserver is a tool that provide BLAST facility. In this database, we have taken care of pan-genomic perspective of large genome and provided group wise BLAST databases. For example, if a user wants to do a BLAST search against only A genome of all the wheat varieties, he/she can select ‘Group A’ and easily do the search.
Kablammo accepts BLAST result output files in XML format and will illustrate exactly which portions of the query sequence mapped to which portions of the subject sequence. Kablammo takes an XML file as an input (which can be downloaded from SequenceServer output) and provides an interactive figure for each subject matched. The query as well as subject sequences can be zoomed-out or zoomed-in for better results viewing experience. Other features like image export, alignment viewer, and exporter are also available.
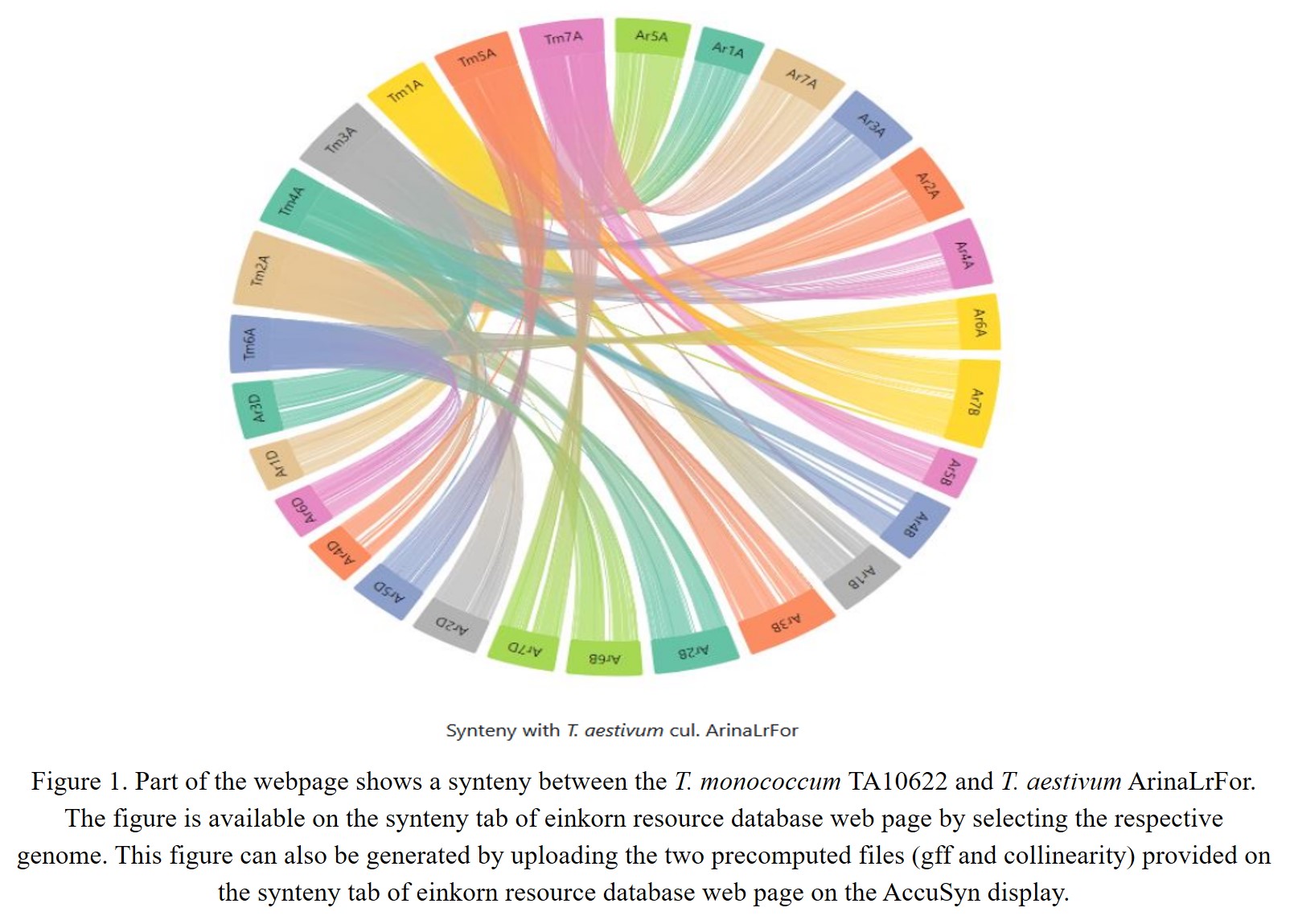
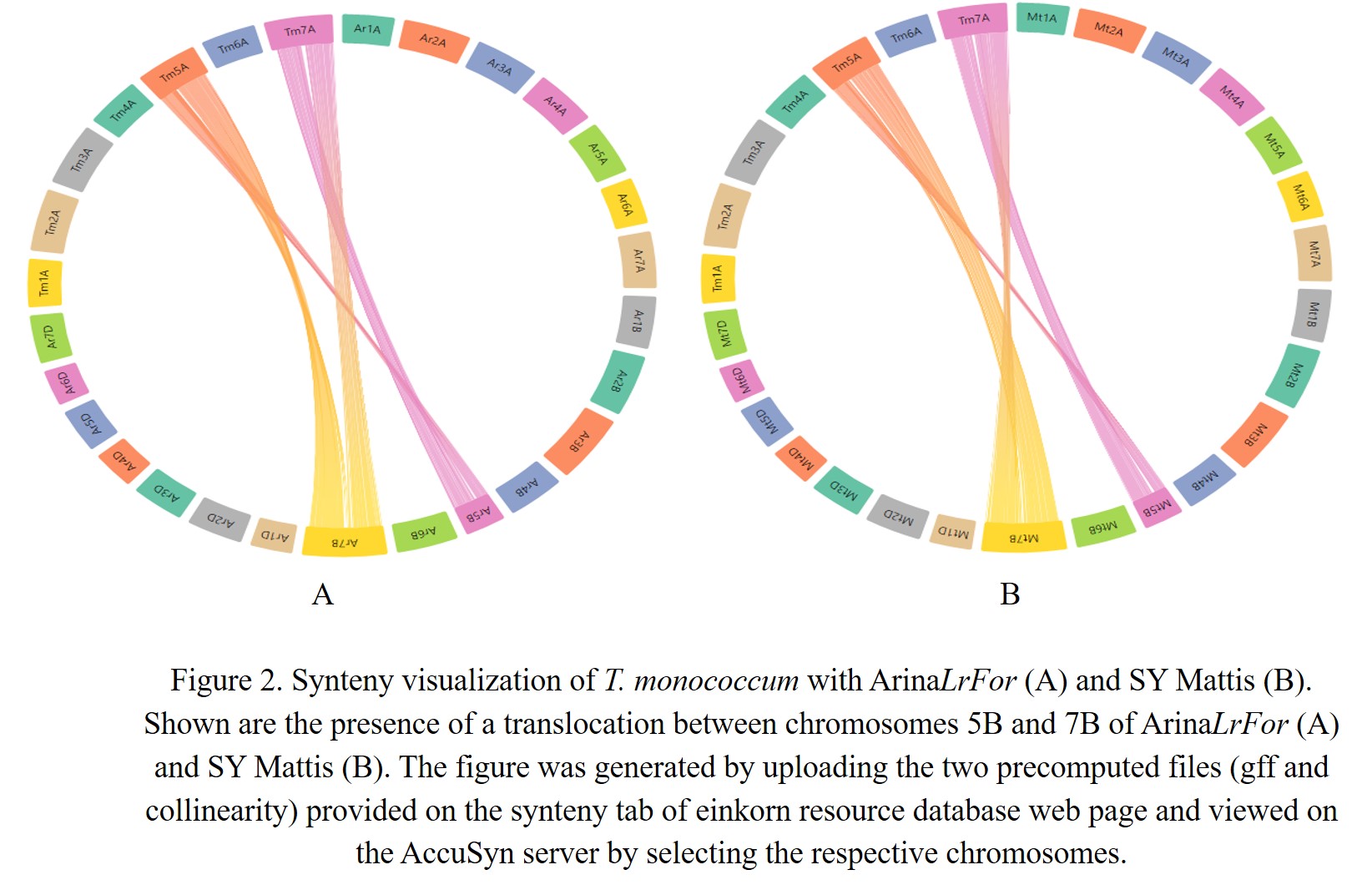
In the present database, we have provided pre-computed synteny results for 29 wheat varieties with the domesticated T. monococcum accession TA10622. These results can be viewed by selecting the relevant variety from the dropdown menu on the left side of the synteny tab on the webpage. Users can have an overview of the synteny between the TA10622 and other wheat varieties. Figure 1 shows synteny between T. monococcum TA10622 and T. aestivum ArinaLrFor and AccuSyn (Jorge Núñez et al., 2020) is interactive software that shows circular syntenic plots of chromosomes and draws links between similar blocks of genes using Simulated Annealing to minimize link crossings. This tool requires two input files viz. an annotation file (.gff) and an alignment file generated from McScanX (Y. P. Wang et al., 2012). We have therefore provided the input files (GFF and collinearity files) to be uploaded into AccuSyn. Users can download these zipped files for different wheat varieties from the dropdown list provided on the right side of the synteny tab. Then they need to extract and upload the two files to the Accusyn website, for which a link has been provided on the same tab. AccuSyn displays synteny between genomes by circularly displaying chromosomes. A user can select one or more chromosomes as required. An interactive figure on the right displays the blocks that were matched between the chromosomes. It should be noted that the same input files can be used as input in SynVisio (https://synvisio.github.io) which can display synteny in a linear fashion which is useful for viewing a few chromosomes at a time. To assess the practical applicability of this synteny tool we took an example from the recent bread wheat pan-genome paper (Walkowiak et al., 2020). In this paper, the authors studied the translocation between chromosomes 5B and 7B in bread wheat lines ArinaLrFor and SY Mattis. As our synteny analysis maps/relates the T. monococcum A genome to all the A, B, and D genomes of hexaploid wheat, we wanted to look into the aforementioned 5B/7B translocation through our synteny tool. For this, the zipped files provided for ArinaLrFor and SY Mattis were used for visualization purposes. Figure 2A shows the synteny view of ArinaLrFor chromosomes 5B and 7B with T. monococcum chromosome 5A. This shows the presence of a translocation between ArinaLrFor chromosome 5B and 7B through our synteny analysis. The same phenomenon was also observed with wheat variety SY Mattis chromosome 5B and 7B (Figure 2B).
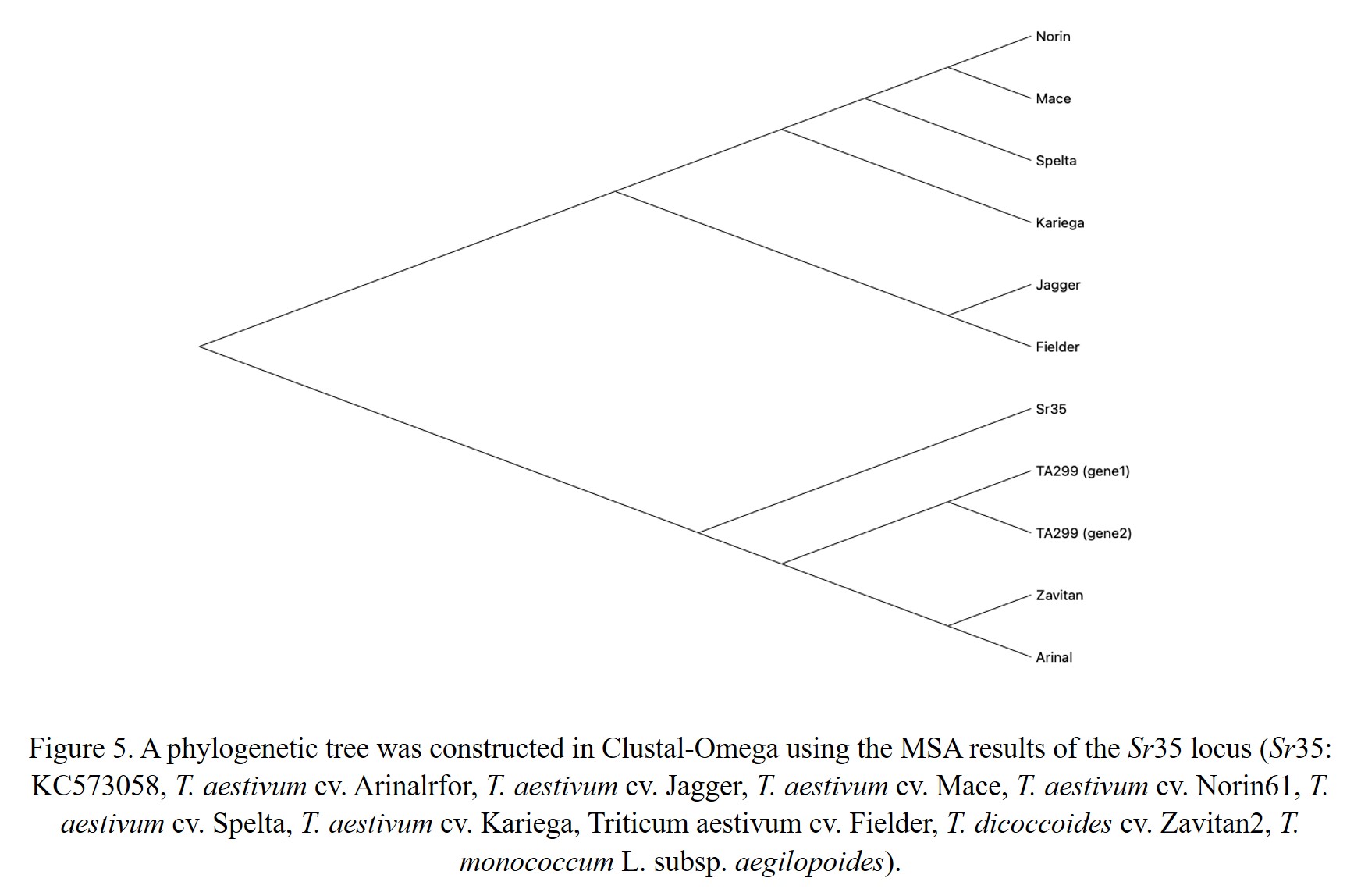
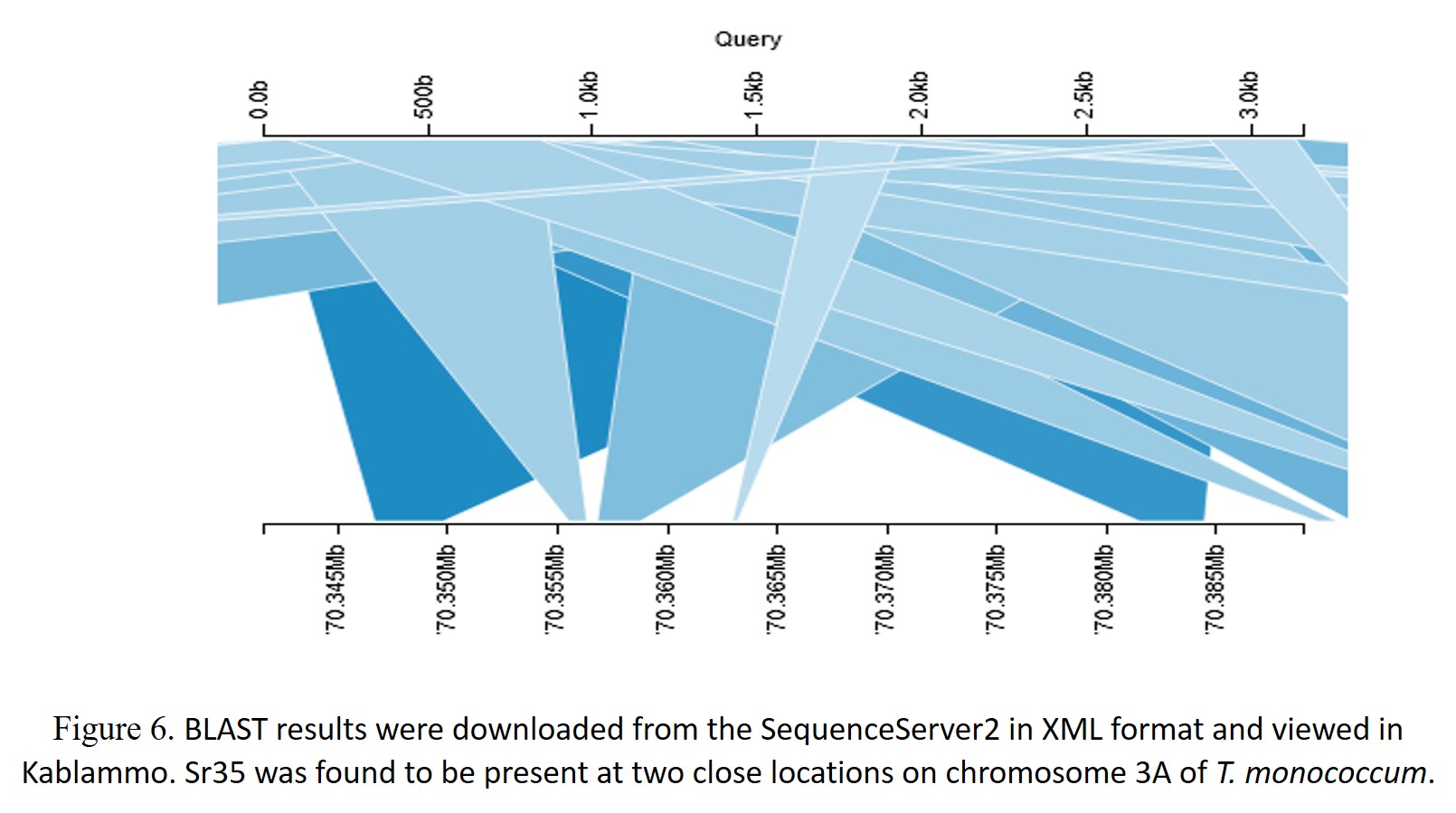
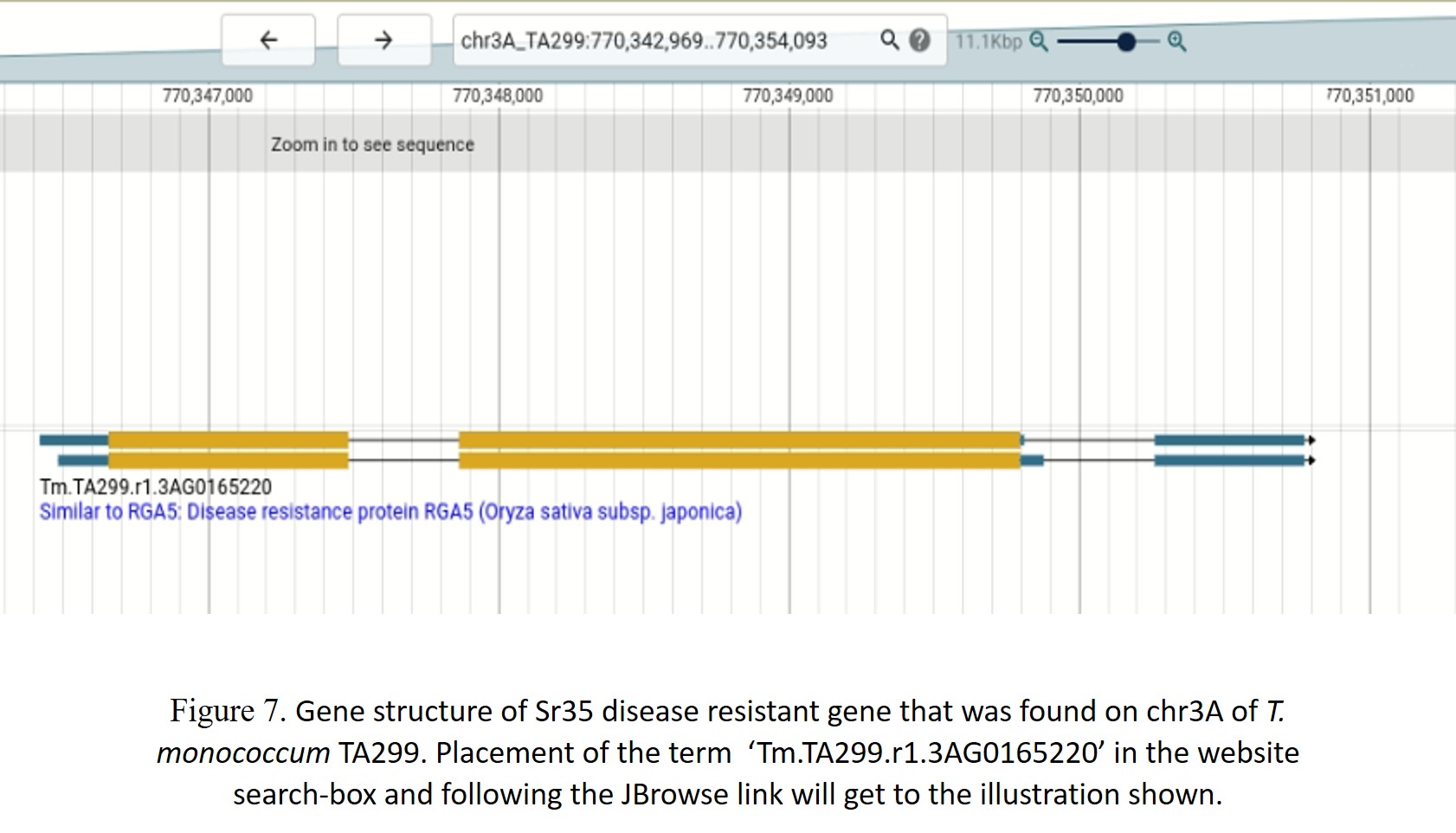
Sr35 is the first gene cloned for stem rust resistance, encoding an intracellular immune receptor of the nucleotide binding site – leucine-rich repeat (NLR) protein family (Saintenac et al., 2013). This gene originates from T. monococcum chromosome 3A. The gene sequence (3151 bp) of Sr35 was BLAST searched against the wheat pangenome using a chromosome group wise search. Hits were seen in all the chromosome groups except group 4. Significant matches with high identity were found on chromosome 3 in all the 57 genome entities matched (Figure 3). However, screening each alignment yielded unique sequences/patterns in different cultivars. This information was particularly useful in marker-assisted selection for stem rust. Filtering the blast results with sequence identity >90% and query coverage >95% (from Group3 hits), removed most of the non-significant hits, including the B sub-genome matches. Significant blast hits on A-genome consisted of genomes i.e. T. monococcum L. subsp. aegilopoides (TA299), tetraploid wheat accession Zaviton, and wheat cv. ArinaLrFor, Norin, Mattis, Mace, Jagger, Julius, Landmark, Kariega, and T. spelta, after removal of D-genome counterparts. For sequence comparison, we extracted 500bp extended sequences from the respective cultivars. The sequence identity ranges from 91 to 96% between the Sr35 and wheat cultivars. Figure 4 shows the part of the multiple sequence alignment of the Sr35 coding gene with the selected genomes. An insertion in the Fielder and ArinalLrFor genome was observed. The phylogenetic tree showed two distinct groups (Figure 5). These two groups differed in terms of SNP and Indel. To demonstrate the similarity at the protein level, we performed gene prediction in the regions, which showed wide differences in the gene structure, with the number of predicted exons ranging from 1 to 4, making these targets as pseudo genes with respect to Sr35 reference. Sr35 seems to have a unique introgression, with missing or pseudo-gene homologs between the cultivars. A close inspection in the case of T. monococcum TA299 helped to reveal two significant matches (Figure 6) on chromosome 3A. Both the two matches were within a 30kb region at position 770.4Mb. We then used JBrowse2 to look into the corresponding positions on the assembly with annotations. Both the two regions matched have only a single type of disease-resistant gene i.e. “Disease resistance protein RGA5” (Figure 7). However, it is interesting to note that around this position there are a lot of other insignificant matches were found which again supports the presence of pseudogenes around this position.


All Rights Reserved. Prior permission required for data duplication.