Currently, the database is searchable using the 'gene Id' of 3 reference assemblies i.e. A. tauschii TA10171, A. tauschii TA1675 and A. tauschii TA2576. As an example, searching for the gene id 'Aet.TA10171.r1.1D000010' will show the respective gene information. The resultant table will show the accession, chromosome number, Gene ID, Gene Ontology (GO), start and end position, and the DNA strand. Links have been provided in the table to view the gene in JBrowse2.
Linear genome viewer of JBrowse2 has been used as a genome browser to browse the genome assemblies of A tauschii acc. TA10171 (Lineage 1), A. tauschii acc. TA1675 (Lineage 2) and A tauschii acc. TA2576 (Lineage 3). All the three assemblies can be accessed by individual chromosomes. Gene model annotations were linked to each of the assemblies which include gene structure visualization for both the High and Low confidence genes. Linear synteny can be viewed between all the three lineages and Chinese Spring
Linear synteny has been provided between the available genomes (currently 4) in the browser. Steps to view the linear synteny is as follows:
1. From the JBrowse2 home page click on Empty link (highlighted).
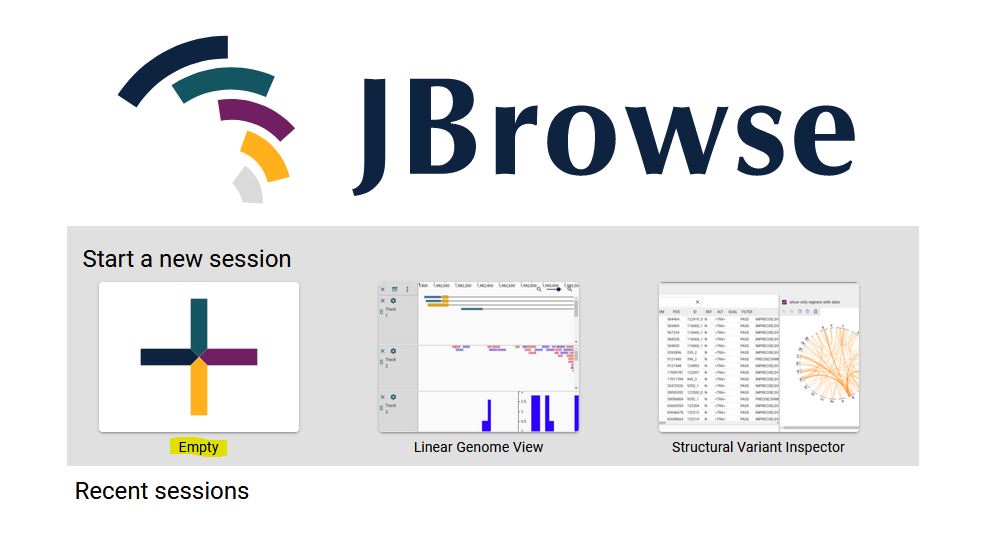
2. Select Linear synteny view from the dropdown menu and click ‘Launch view’.
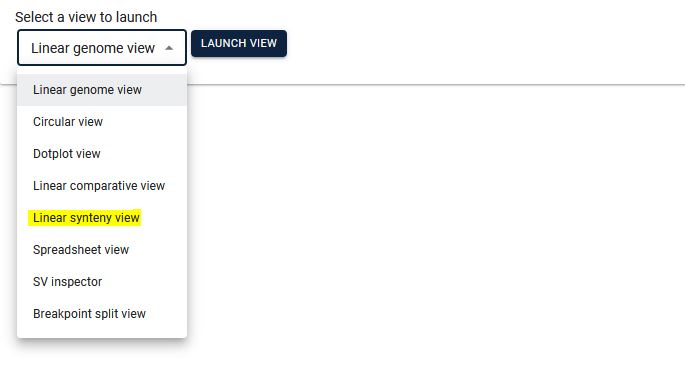
3. Select the two species for which you want to see synteny, then select the existing track (automatically the respective file will be displayed) and click Launch.
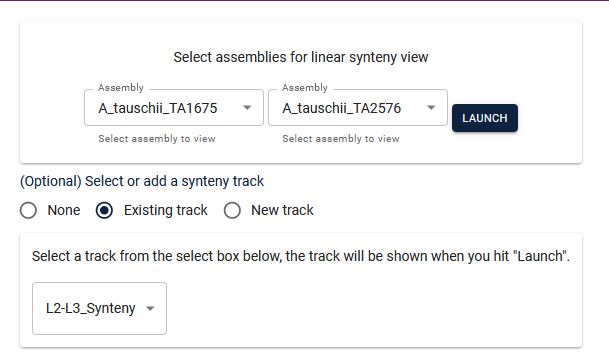
4. From the two dropdown menus, select the respective chromosomes (it will be best to choose the same chromosome number on both the genomes).
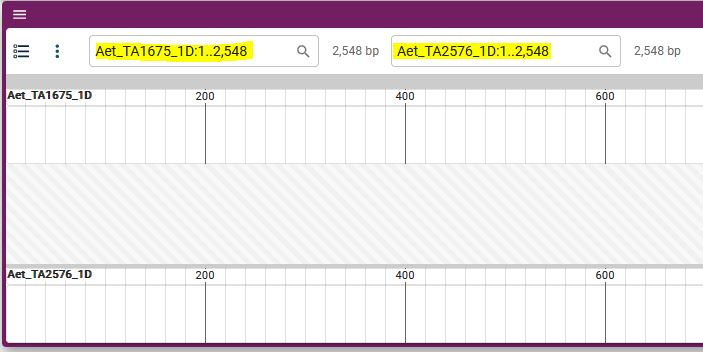
5. Synteny will be displayed.

Sequenceserver is a tool that provides BLAST facility. In this database, we have added the three reference A. tauschii reference assemblies, against which a BLAST search can be performed.
Kablammo takes an BLAST XML file as an input (which can be downloaded from SequenceServer output) and provides an interactive figure for each subject matched to the query. The query as well as subject sequences can be zoomed-out or zoomed-in for better results viewing experience. Other features like image export, alignment viewer and exporter are also available.
In the present database, we have provided pre-computed synteny results for 25 wheat varieties compared with all the three tauschii reference assemblies from three lineages. These results can be viewed by selecting the relevant varieties from the dropdown menus on the left side of the synteny tab on the webpage. Here, users can have an overview of the synteny between the selected wheat varieties. AccuSyn (Jorge Núñez et al., 2020) is an interactive software that shows circular syntenic plots of chromosomes and draws links between similar blocks of genes using Simulated Annealing to minimize link crossings. This tool requires two input files viz. an annotation file (.gff) and an alignment file generated from McScanX (Y. P. Wang et al., 2012) (.collinearity). We have therefore provided the input files (GFF and collinearity files) on our webpage, that can be downloaded and uploaded at AccuSyn server. Users can download these zipped files for different wheat varieties from the dropdown list provided on the right side of the synteny tab. Then they need to extract and upload the two files to the Accusyn server, for which a link has been provided on the same tab. AccuSyn displays synteny between genomes by circularly displaying chromosomes. A user can select one or more chromosomes as required. An interactive figure on the right displays the blocks that were matched between the chromosomes. It should be noted that the same input files can be used as input in SynVisio (https://synvisio.github.io) which can display synteny in a linear fashion which is useful for viewing a few chromosomes at a time.
All Rights Reserved. Prior permission required for data duplication.